| Programmer: Wade
Lo Art designers: SY Cheng & Jong Liu Lab of Dr. Ping-Chiang Lyu, Institute of Bioinformatics and Structural Biology, NTHU, Taiwan, R.O.C. |
Introduction
User Interface
Search by specifying organism and metabolic pathway
Search by keyword for organism
Gain further information about KEGG non-recorded enzymes
Resources
KPST is a lite search engine for KEGG pathway database. Users can do the search by specifying organism or metabolic pathway; besides, a key word search from organism is also available. KPST is featured by it's capability to make clickable not only discovered (KEGG recorded) but also undiscovered (KEGG non-recorded) enzymes from a pathway map for users to look for further information about them.
This tool is built in Nov., 2004 and last updated in Jan., 2006 by Wade Lo. Suggestions are welcomed and can be mailed to d914226@oz.nthu.edu.tw.
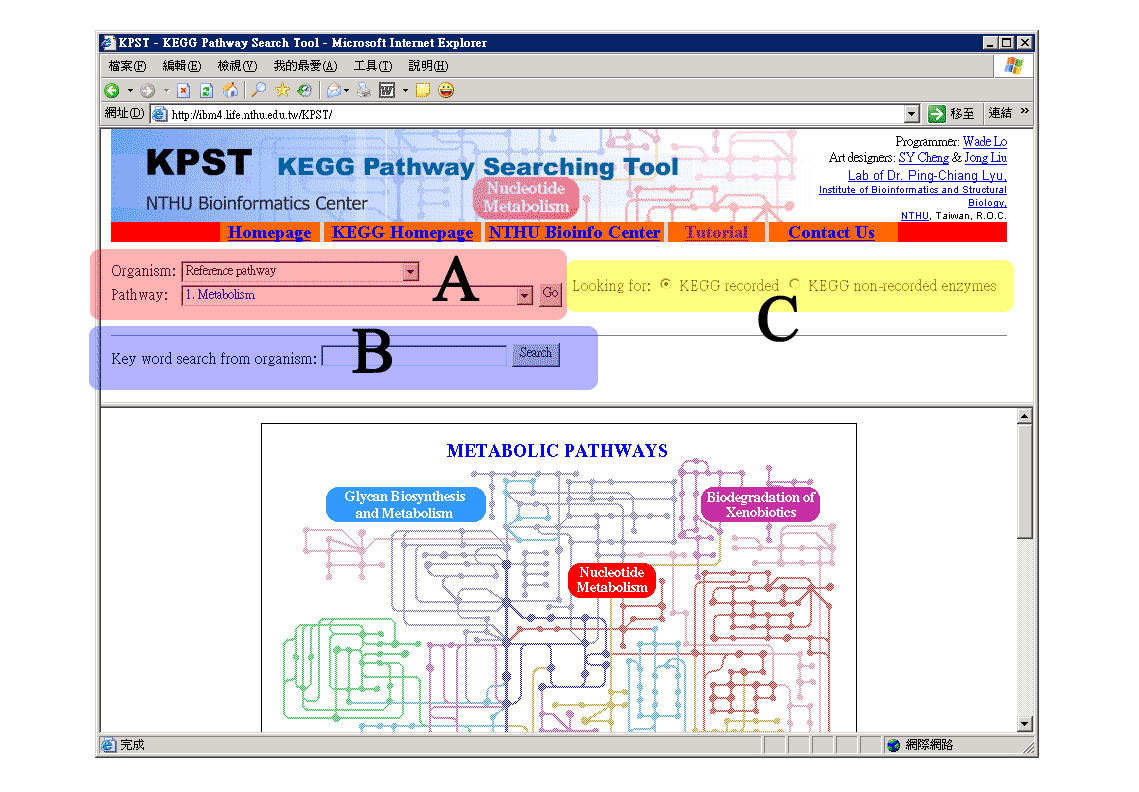
The user interface of KPST is divided into two domins (frames). The upper one is the operational domain while the lower domain is where the search results come from. There are three functional parts of the upper domain (e.g. the A, B, C region in the following figure) and they can be used to:
A. Search by specifying organism and metabolic pathway.
B. Search by keyword for organism.
C. Gain further information about KEGG non-recorded enzymes.
 |
|
Search by specifying organism and metabolic pathway
Region A is used for specific search. One can finds unique information by specifying organism and metabolic pathway stepwise as follows:
 |
These are the original options. | |
| |
||
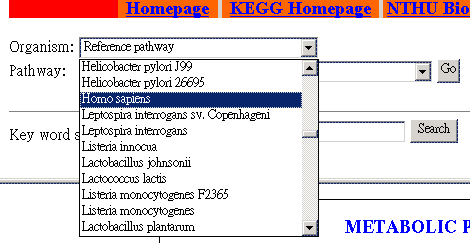 |
Select an organism. | |
| |
||
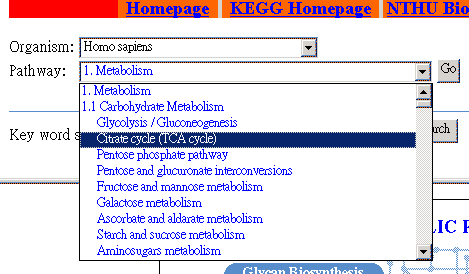 |
Select a metabolic pathway. | |
| |
||
 |
After we click "Go", the result like this will be displayed in the lower domain of KPST webpage. |
Search by keyword for organism
If someone is not sure about the full name of his interested organism, the keyword search may offer some help:
Gain further information about KEGG non-recorded enzymes
http://www.genome.jp/dbget-bin/get_pathway?org_name=xac&mapno=00052
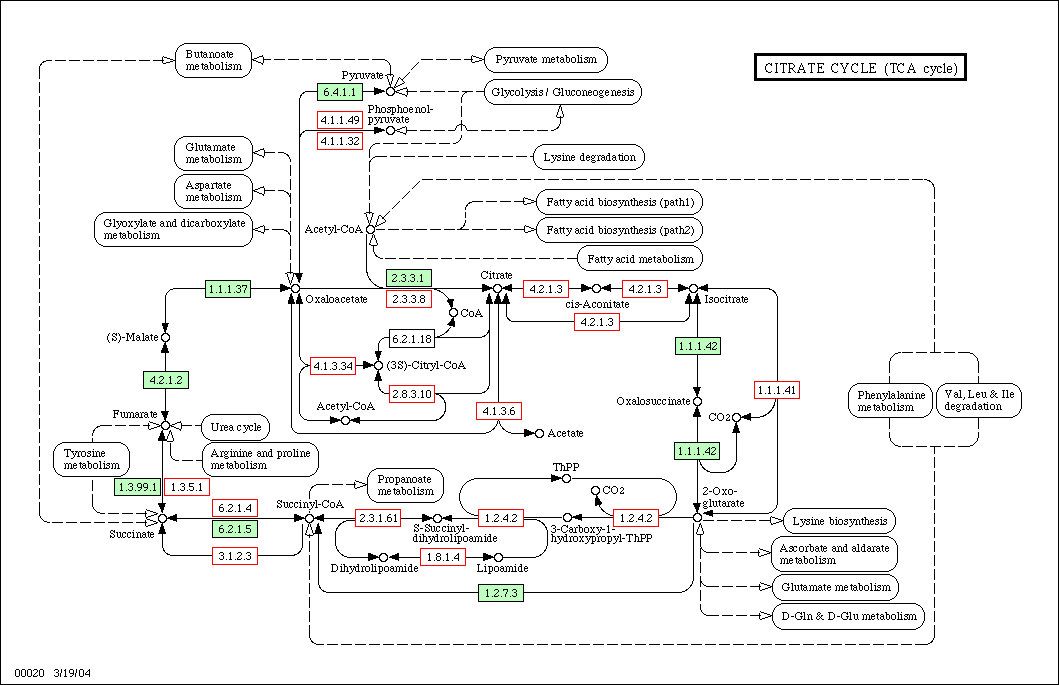
In a traditional result page of KEGG website just like what the above hyperlink can give you, green boxes are used to mark the enyzmes which have been discovered or identified, whereas the enzymes in white boxed are not. Only the green boxes are clickable for further information.
KPST can analyze the result image of KEGG and then make the white boxes marked enzymes clickable for further information from swissprot ENZYME database. One can enable this feature simply by changing the selection in functional region C to "KEGG non-recorded enzymes."
 |
If this feature has been enabled, after we do the search, the orginally white-box-marked enzymes would now be marked by clickable red boxes:
KEGG - http://www.genome.jp/kegg/kegg.html
KEGG2 - http://www.genome.jp/kegg/kegg2.html
swissprot ENZYME - http://au.expasy.org/enzyme/